The Joint Open Genome and Omics Platform (JoGo), the world's first database containing haplotypes covering all human genes, has been released
- Others
- Funding
- Database Integration Coordination Program
On Nov 29, 2025, Professor Masaaki Nagasaki at Medical Institute of Bioregulation, Kyushu University and his colleagues announced the release of the Joint Open Genome and Omics Platform (JoGo), a database containing a comprehensive haplotype catalog of human genes, in a special database issue of the scientific journal "Nucleic Acids Research." This article was selected as a breakthrough article in this issue. Kyushu University, the RIKEN Institute, the National Center for Global Health and Medicine (NCGM), the Information and Systems Research Organization's Database Center for Life Sciences (DBCLS), Kitasato University, and the Japan Science and Technology Agency (JST) issued a press release on Dec 3, 2025, outlining the details and functionality of the data included in JoGo, as well as its features.
Previous genetic mutation analysis has focused on investigating the effects of individual gene mutations or polymorphisms. However, the effects of genetic mutations or polymorphisms, such as constitution, can sometimes be determined by the combination of multiple genetic mutations and polymorphisms (haplotypes (*1)). A familiar example of this is the ABO blood type (*2).
Haplotype analysis has been performed only on limited gene groups so far, such as the cytochrome P450 (CYP) gene group involved in drug metabolism, and has been used for clinical applications such as classifying drug metabolism capacity and optimizing drug administration. While the importance of haplotype analysis has long been recognized, it was difficult to determine accurate haplotypes across the entire genome using the genome sequences determined by conventional short-read sequencing methods (*3). Consequently, no haplotype database capable of analyzing the entire genome has existed until now.
In recent years, technological advances in long-read sequencing methods (*4) have enabled the determination of human genome sequences with accurate haplotypes across the entire genome. Consequently, Professor Nagasaki and his colleagues defined haplotypes using the ACTG hierarchical nomenclature (*5): A (variants in the protein coding region involving amino acid substitutions)/C (variants in the protein coding region not involving amino acid substitutions)/ T (variants confined to exonic non-coding sequence)/G (variants in introns within gene region). They also constructed a haplotype catalog using genome sequences from 258 individuals across five continents determined by long-read sequencing, and have released a database containing these haplotypes, the Joint Open Genome and Omics Platform (JoGo). This is the world's first database serving as a genome-wide haplotype catalog. It includes genome sequences from samples originating from 105 Japanese individuals.
Furthermore, Professor Nagasaki and his colleagues have enabled the investigation of relationships between haplotypes defined by the "ACTG hierarchical nomenclature" and gene expression levels in the JoGo Platform. This was achieved by examining gene expression data from 1,280 individuals studied in immortalized B cells across three research projects, along with correlations between haplotypes and gene expression levels in the same samples.
The human genome is considered to be 99.9% identical, but it is thought that the remaining 0.1% of differences determine individual variations in constitution, such as susceptibility to specific diseases or a higher likelihood of experiencing side effects from certain medications. Professor Nagasaki states that by conceptualizing these differences in individual constitution--which could not be explained by variations in individual genes or polymorphisms (i.e., "points")--as haplotypes (i.e., "lines"), which are combinations of sequences, it becomes possible to broadly explain human genetic diversity.
For more details, please refer to the paper "JoGo 1.0: the ACTG hierarchical nomenclature and database covering 4.7 million haplotypes across 19,194 human genes" and the press release.
<Number of Data Included in JoGo Platform v.1.0>(As of Nov 29, 2025)
- Samples Analyzed: 258
(Of which, 105 were from Japanese individuals) - MANE Standard Protein-Coding Genes: 19,194
- Haplotypes: 4,656,478
A (variants in the protein coding region involving amino acid substitutions): 174,376
C (variants in the protein coding region not involving amino acid substitutions): 300,610
T (variants confined to exonic non-coding sequence): 486,288
G (variants in introns within gene region): 3,695,204
JoGo Platform is developed as a part of JST Database Integration Coordination Program (DICP), "Development of research and educational platform with open human genomic and omics international database" (Principal Investigator: NAGASAKI Masao, Professor, Medical Institute of Bioregulation, Kyushu University).
Terminology
*1 Haplotype: A combination of multiple changes (mutations) inherited together on the same chromosome. These mutations can affect function more collectively than individual mutations.
*2 ABO blood type: ABO blood types are determined by the combination of two types of sugar chain antigens on the surface of red blood cells. Type A sugar chains have N-acetylgalactosamine and type B sugar chains have galactose on their terminals, respectively. Differences in the combination of sequences of two glycosyltransferase gene loci on two chromosome 9 that make type A and type B sugar chain inherited from both parents result in the differences AA, AO, BB, BO, AB, and OO ("O" means that the sugar chain of that type cannot be attached), and blood types are divided into A (AA, AO), B (BB, BO), AB (AB), and O (OO).
*3 Short-read sequencing: A sequencing method using equipments generally known as a "next-generation sequencers". It has been used to determine the majority of human genome sequences. It is a base sequence determination method in which DNA is digested into short fragments, and short sequences of approximately 100-150 bases at both ends are read simultaneously in parallel in large quantities, and these short sequences are then assembled on a computer. While this method allows for low cost and suitable for large-scale analysis, it cannot detect repeted sequence regions, or large structural variations such as insertions, deletions, translocations, and inversions that exceed the read length.
*4 Long-read sequencing: A method of determining base sequences that reads the sequence of long DNA fragments of several thousand to tens of thousands of bases at a time. Unlike short-read sequencing (*3), it can detect repeted sequence regions and large structural variations, and can distinguish and sequence the genomes inherited from each parent. Therefore, it is a genome sequence that allows for haplotype analysis, which was difficult with genome sequences determined by short-read sequencing.
*5 ACTG hierarchical nomenclature: A new haplotype nomenclature method proposed in this study that can be applied genome-wide. Gene sequence types are represented in the hierarchy of A (variants in the protein coding region involving amino acid substitutions), C (variants in the protein coding region not involving amino acid substitutions), T (variants confined to exonic non-coding sequence), and G (variants in introns within gene region), and IDs are assigned in order of frequency.
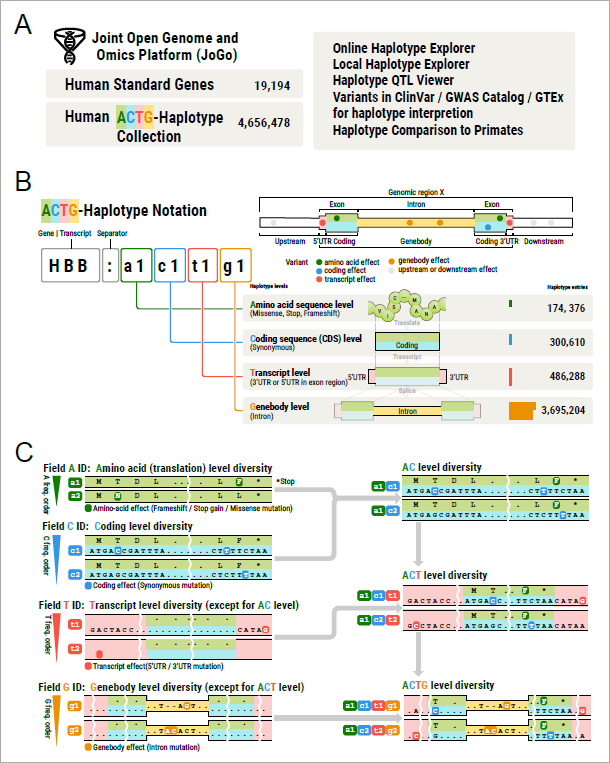
Figure 1. (A) Overview of JoGo portal contents and functions. (B) Overview of the ACTG-haplotype notation and the haplotype collections in the JoGo database. (C) Example of A-, C-, T-, and G-level haplotype ID assignment and hierarchical haplotype ID construction.
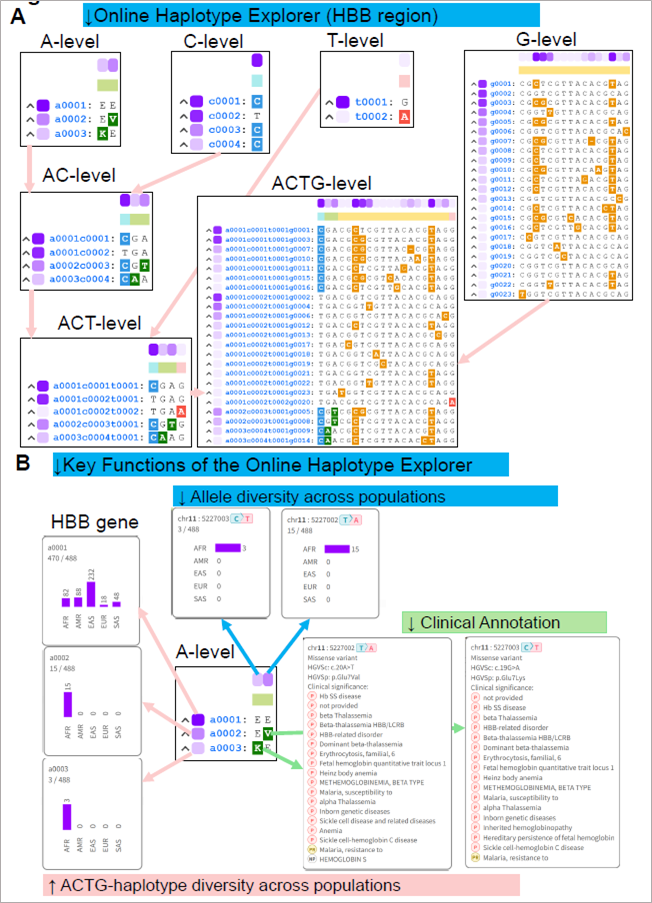
Figure 2. (A) Online Haplotype Explorer view for the HBB locus, illustrating hierarchical ACTG-haplotype notation from the A-level through the C-level, T-level, and G-level, as well as combined AC-, ACT-, and full ACTG-level haplotype structures. Insets show ranked haplotype IDs (e.g., a1, c1, t1, g1) with representative sequence motifs and color-coded allele differences. (B) Detailed view of the Online Haplotype Explorer for A-level haplotypes at the HBB locus. The top color bar encodes the global frequency of each variant (darker shading indicates higher frequency), with hover-activated bar-plot tooltips displaying allele counts across JoGo reference populations. The left color bar encodes the global frequency of each A-level haplotype, with similar tooltips for haplotype counts. Clicking or hovering on a variant also reveals ClinVar annotations and provides a direct link to the corresponding record in TogoVar, a companion database to JoGo that aggregates variant annotations.
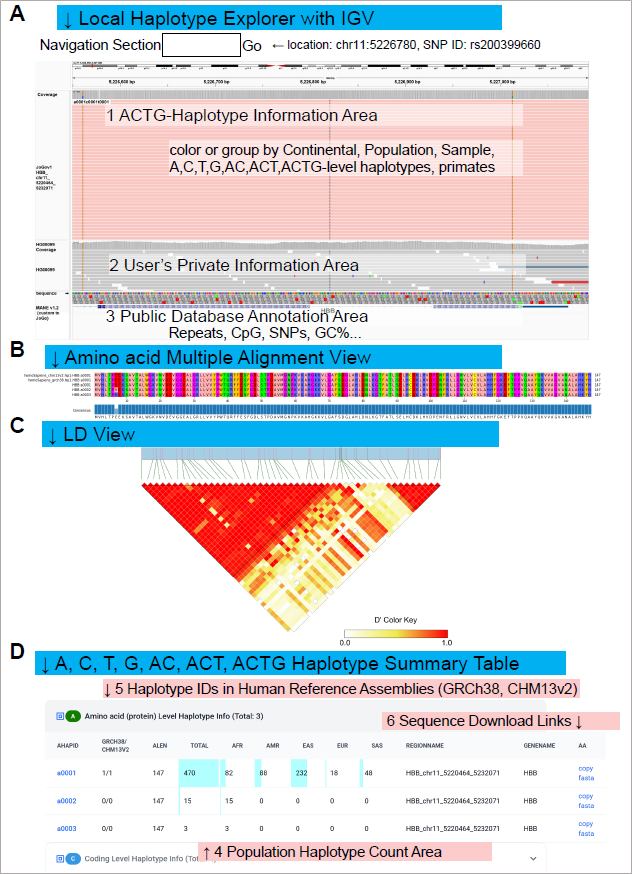
Figure 3. (A) Local Haplotype Explorer session for the HBB locus in IGV. JoGo's per-gene ACTG-haplotype dictionary is provided as a pre-aligned BAM, with each "read" representing a haplotype and custom tags for A, C, T, G (and AC, ACT, ACTG) IDs, as well as population labels. Public JoGo reference tracks (region 1) and the same public JoGo data loaded privately into IGV (region 2) are displayed together on the GRCh38 coordinate (chr11:5,226,550-5,227,092), enabling secure, side-by-side exploration of shared versus divergent sequence tracts. (B) Multiple-sequence alignment of deduced A-level protein haplotypes for HBB, including the GRCh38 and CHM13v2 reference coding sequences. (C) ACTG-level linkage-disequilibrium (LD) heatmap for the HBB locus. Cells are shaded by pairwise D′ values between coding variants (dark = strong LD; light = weak LD), indicating haplotype structure at the nucleotide level. (D) Population-specific counts of A-level HBB haplotypes across the five JoGo reference populations (EAS, AFR, AMR, SAS, EUR). Bars indicate the number of distinct A-level haplotypes observed in each population, allowing for a comparison of haplotype diversity within and between populations.
Related Links
- Press Release: "Comprehensive Human Gene Types: JoGo Database Open to the Public - Contains 4.7 Million Sequence Types for 19,000 Genes, Boosting Disease and Genetic Analysis" (Dec 3, 2025) | JST
- "JoGo 1.0: The ACTG Hierarchical Nomenclature and Database Covering 4.7 Million Haplotypes Across 19,194 Human Genes" | Nucleic Acids Research
- Database: JoGo Platform
- Development of research and educational platform with open human genomic and omics international database | NBDC
Project summaries and reports are posted.