A paper was published on ProteomeXchange, the international consortium for proteomics resources
- Others
- Funding
- Database Integration Coordination Program
On Nov 11, 2025, a paper about ProteomeXchange, an international consortium of proteomics resources, was published in the database Issue of the scientific journal Nucleic Acids Research. The paper also mentioned the work achieved by jPOST, which is developed by Professor Yasushi Ishihama of Kyoto University, who is also a co-author of this paper.
The ProteomeXchange consortium was established in 2012 to standardize open data management in mass spectrometry-based proteomics. Currently, major proteomics data repositories around the world, including PRIDE (EU), MassIVE (USA), jPOST (Japan), and iProX (China), as well as smaller proteomics data resources, Panorama Public (USA) and PASSEL (USA), work together under the ProteomeXchange umbrella. Based on the FAIR Principles for biological data, ProteomeXchange operates a common web portal, ProteomeCentral, to access all datasets published by its affiliated data repositories. As of Jun 2025, 64,330 datasets have been deposited in the ProteomeXchange resource. In Dec 2022, ProteomeXchange resources were selected as Global Core Biodata Resources by Global Biodata Coalition, recognizing them as essential biological resources for the scientific community.
This paper describes ProteomeXchange's efforts to standardize proteome data formats and promote the reuse of proteome datasets. It also introduces jPOST's unique initiatives, including the publication of the proteomics data journal "Journal of Proteome Data and Methods (JPDM)," which encourages researchers to register data in a reusable format on the ProteomeXchange resource, and the development of "UniScore," a unified standard scale developed to overcome differences in the measurement environments of various samples and enable comparative quantitative analysis between samples.
For more details, please see the paper "The ProteomeXchange consortium in 2026: making proteomics data FAIR."
jPOST consists of 1) "jPOSTrepo", a repository of proteome mass spectrometry data, 2) a proteome raw data reanalysis system based on standardized protocols, and 3) "jPOSTdb", a database integrating the reanalysis data. jPOST is developed as a part of JST Database Integration Coordination Program (DICP), "jPOST prime: proteome database environment based on community cooperation " (Principal Investigater: ISHIHAMA Yasushi, Professor, Graduate School of Pharmaceutical Sciences, Kyoto University).
(Figure) The ProteomeXchange consortium initiatives
Members of The ProteomeXchange consortium: PRIDE (EU), MassIVE (USA), jPOST (Japan), and iProX (China), Panorama Public (USA) and PASSEL (USA).
(Table) Major proteome mass spectrometry data repositories and their features
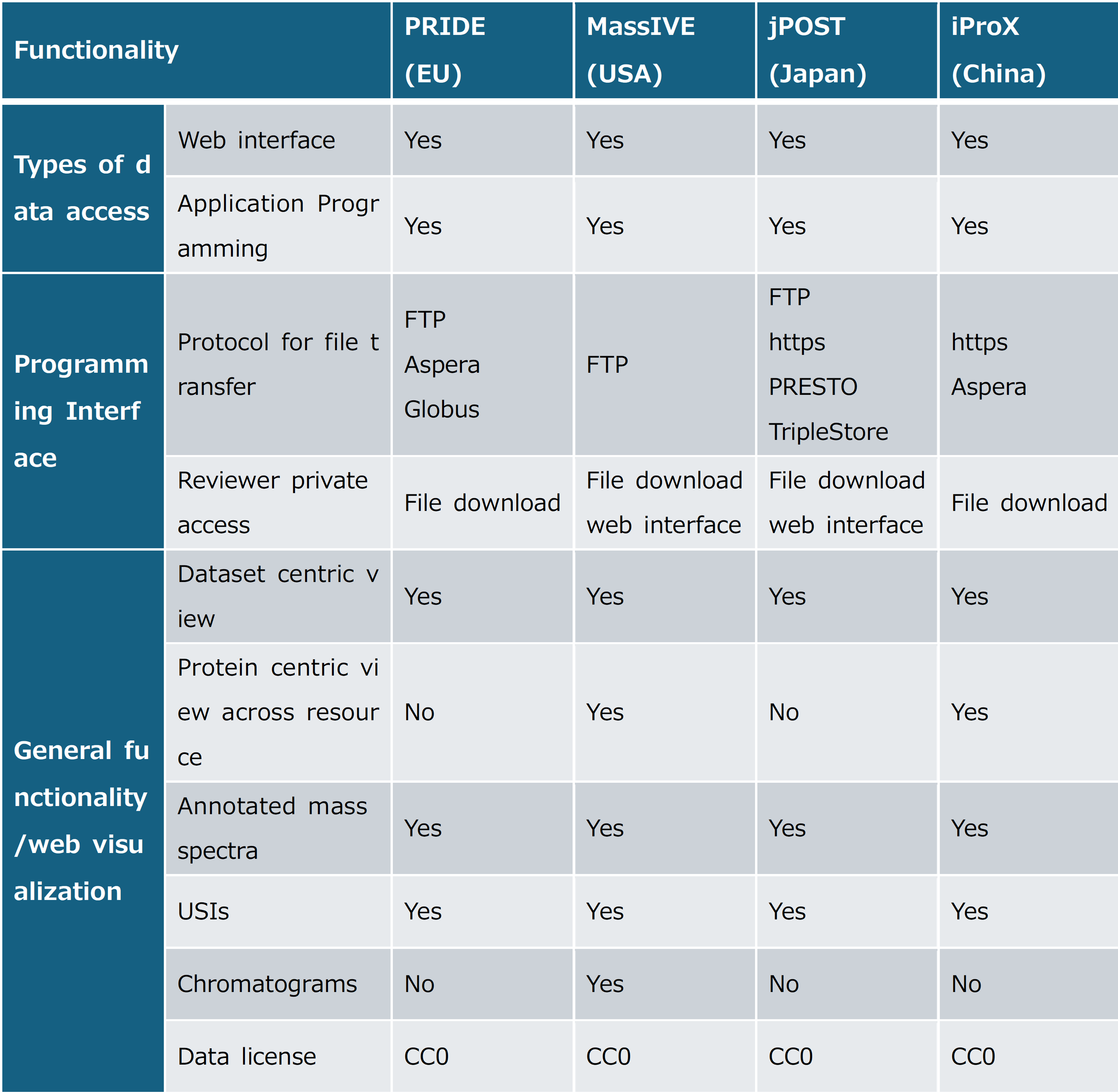
Supplementary information
*FAIR Principles: Guidelines that outline principles for making data "Findable," "Accessible," "Interoperable," and "Reusable."