Human and mouse cis-element data analyzed by fanta.bio is now available as a resource in FANTOM6
- Others
- Funding
- Database Integration Coordination Program
A paper on the cis-element database "fanta.bio" developed and provided by KASUKAWA Takeya, Team Leader, Center for Integrative Medical Sciences, RIKEN, and his colleagues has been published in the Database Issue of the scientific journal "Nucleic Acids Research" on Nov 27, 2024. The paper reports on the expansion of functional information on non-coding RNAs and the development of cis-element data in the international project "FANTOM6", and "fanta.bio " integrates these cis-element data with data from other public databases and makes them publicly available.
A cis-element is a genomic region involved in the transcriptional regulation of a gene. "fanta.bio" contains information on the genomic location of cis-elements, activity information based on expression levels, neighboring genes and single nucleotide polymorphisms (SNPs). This information is based on the human and mouse cis-element regions identified by reanalysis of the FANTOM6 dataset as the core, integrated with data from other public databases. The FANTOM6 dataset was reanalyzed using a newly developed method developed by Deputy Director KAWAJI Hideya and his colleagues at the Tokyo Metropolitan Institute of Medical Science to identify promoters and enhancers from CAGE* data, and 447,315 and 288,877 were identified in human and mouse, respectively. Data being integrated include information on transcription factors that bind to the cis-element from ChIP-Atlas and genomic mutation information from TogoVar and MOG+. These information can be viewed side-by-side using the UCSC Genome Browser.
This article describes the data processing methods, datasets, visualization methods, and offerings described above in fanta.bio, as well as the linkage to external databases. RIKEN, the Tokyo Metropolitan Institute of Medical Science, and JST issued a press release in conjunction with the publication of this paper, stating their expectations for the paper's use in basic research on gene expression regulation mechanisms, disease research, and drug discovery research.
For more details, please see the published article "Update of the FANTOM web resource: enhancement for studying noncoding genomes" and the press release "Latest update of the FANTOM web resource - new data on non Update of the FANTOM web resource: enhancement for studying noncoding genomes".
fanta.bio and ChIP-Atlas are part of INTRARED, the Integrated Transcriptional Analysis Platform Database, and are developed as a part of JST Database Integration Coordination Program (DICP), "Construction of an integrative data platform for transcriptional regulations ".
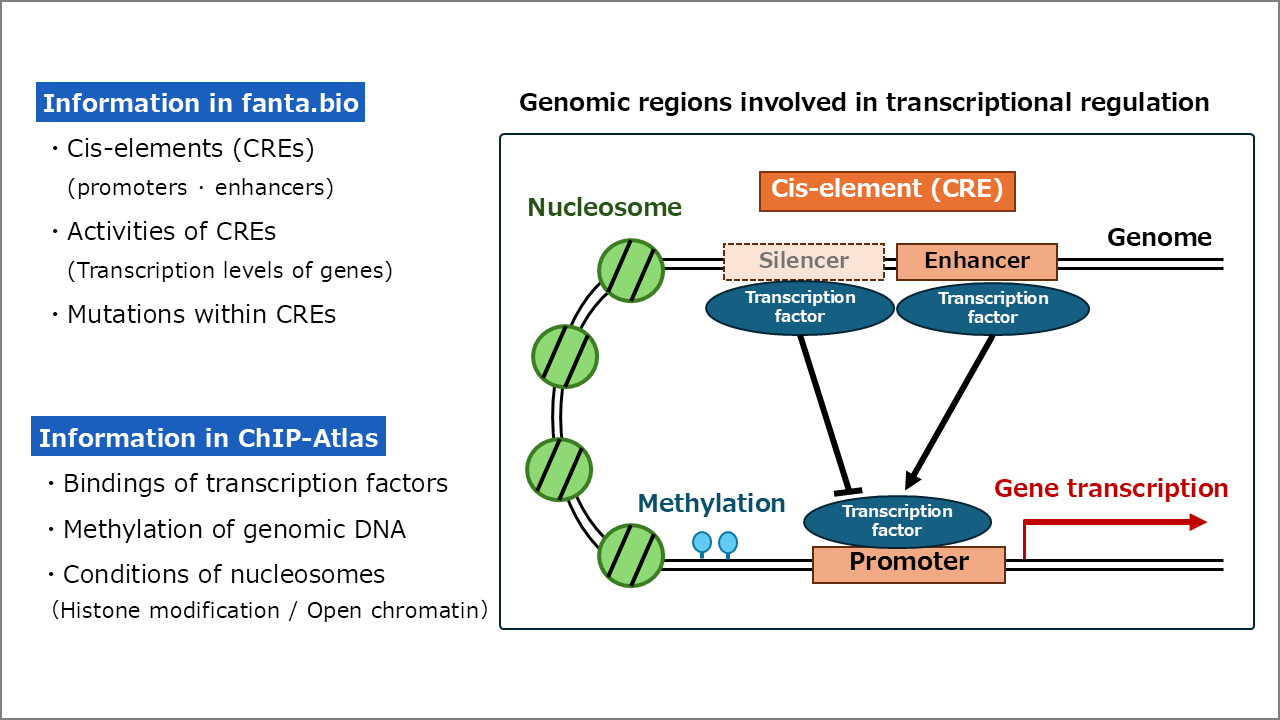
(Figure) Regulators of gene expression: fanta.bio contains information on cis-elements (promoters and enhancers) that control gene expression on the genome, and ChIP-Atlas contains information on transcription factors that bind to these elements, methylation of genomic sequences that control gene expression on/off, and nucleosome status (histone modifications and open chromatin). By integrating and analyzing these information, the mechanism of gene expression regulation can be analyzed.
Term Explanation
FANTOM6: This was the sixth phase of the FANTOM (Functional ANnoTation of Mammalian Genome) Project, an international collaboration led by RIKEN. the FANTOM project has been incorporating new technologies for over 20 years, large-scale transcriptional analysis of mammals, mainly humans and mice, and has provided a variety of data on transcription. CAGE method: An experimental method developed by RIKEN to identify transcription start sites. The enhancer region can be identified by detecting enhancer RNA (eRNA) transcribed in the enhancer region.
Related Links
- "Update of the FANTOM web resource: enhancement for studying noncoding genomes" | Nucleic Acids Research
- Press release "Latest update of the FANTOM web resource - new data on non Update of the FANTOM web resource: enhancement for studying noncoding genomes" (Nov 27, 2024) | RIKEN
- Construction of an integrative data platform for transcriptional regulations | NBDC
Project summary and reports. - FANTOM Project
- INTRARED
- fanta.bio