DeepSpaceDB, a spatial transcriptomics database, is released
- Others
- Funding
- Database Integration Coordination Program
On Sep 4, 2024, VANDENBON Alexis, Associate Professor, Institute for Life and Medical Sciences, Kyoto University, and his colleagues announced the release of DeepSpaceDB, a spatial transcriptomics database.
Spatial transcriptomics is an emerging technology that comprehensively examines which genes are expressed, and at what levels, in different regions of a tissue sample. In recent years, advancements in analytical techniques have led to the accumulation of large amounts of data, which are available in public databases. This data enables us to understand the location and quantity of cells in a tissue, how these cells function, and how changes in these functions are related to diseases at the molecular level. However, the large volume and complexity of spatial transcriptomics data can be challenging for researchers without bioinformatics expertise.
DeepSpaceDB makes it easy to browse spatial transcriptomics data from a wide variety of tissues, and allows users to perform advanced analyses interactively, even without expertise in bioinformatics. The database includes nearly all spatial transcriptomics data of the primary platform, 10x Genomics Visium, that are publicly available from NCBI GEO and other sources. The metadata, such as tissue of origin and disease status of the samples, have been carefully reviewed and standardized in terminology and ontologies. In addition, DeepSpaceDB offers various analysis tools, including clustering of each spot based on expression profiles, visualization of the spatial expression distribution for any gene, display of pathways with spatially variable expression, visualization of the spatial distribution of predicted cell types based on expressed genes, and comparison of gene expression between freely selected areas.
For example, DeepSpaceDB enables researchers to elucidate what types of neurons are present in a brain tissue slice, identify which cells in kidney tissue are affected by renal disease, and find which areas of breast cancer tumors are infiltrated by immune cells, among other applications.
<Number of DeepSpaseDB> (as of Sep 4, 2024)
- Human 626 samples
- Mouse 412 samples
Associate Professor VANDENBON Alexis has presented the official release of DeepSpaceDB in an oral presentation at NGS Expo 2024 (at Osaka International Convention Center on Sep 4 and 5, 2024). He will also present at Symposium of Database Togo (Integration) 2024 "AI + Robotics + Databases Changing Life Sciences" at The Grand Hall Shinagawa on Saturday, Oct 5, 2024, and at the 1st Asia & Pacific Bioinformatics Joint Conference at Naha Cultural Arts Theater NAHArt on Oct 22 to 25, 2024.
DeepSpaceDB is developed as a part of JST Database Integration Coordination Program (DICP), "Development of a database for spatial genomics data analysis (Principal Investigator: VANDENBON Alexis, Associate Professor, Institute for Life and Medical Sciences, Kyoto University)" .
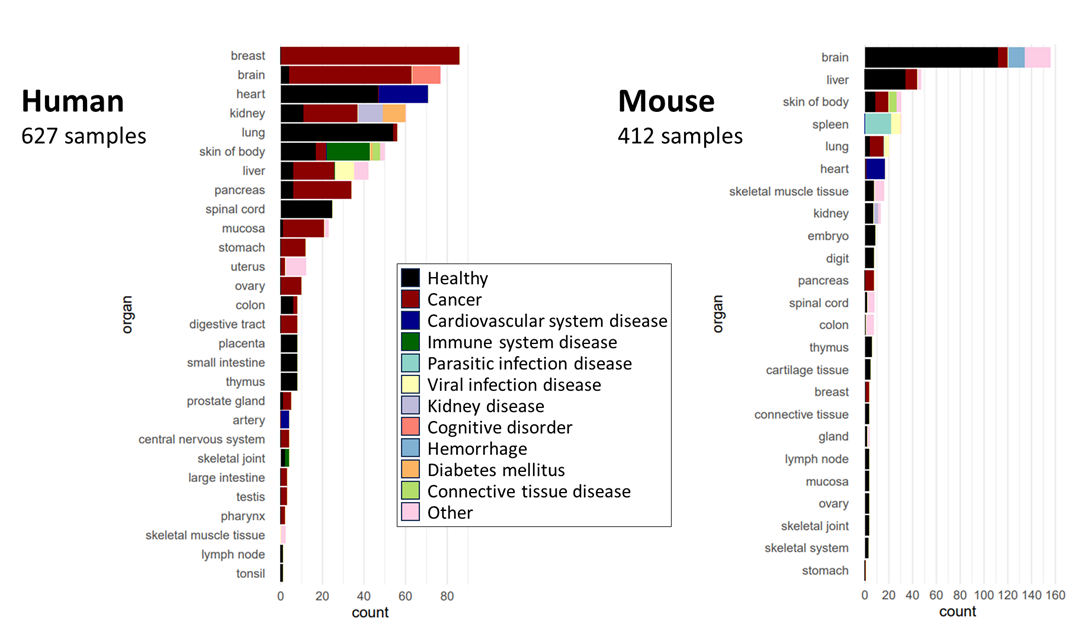
Species and tissue origins of the samples in DeepSpaceDB
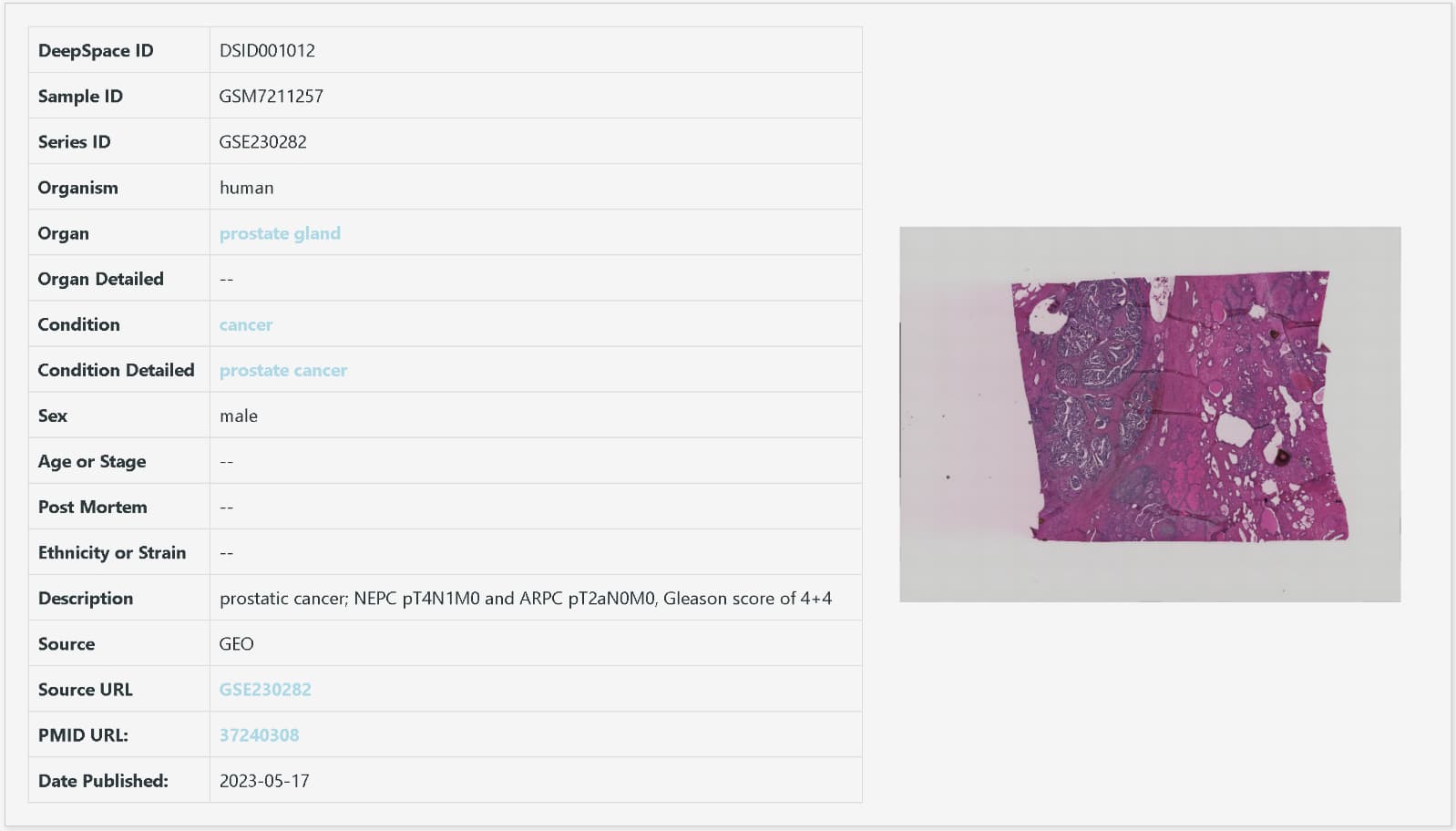
An example of sample metadata (DSID001012)
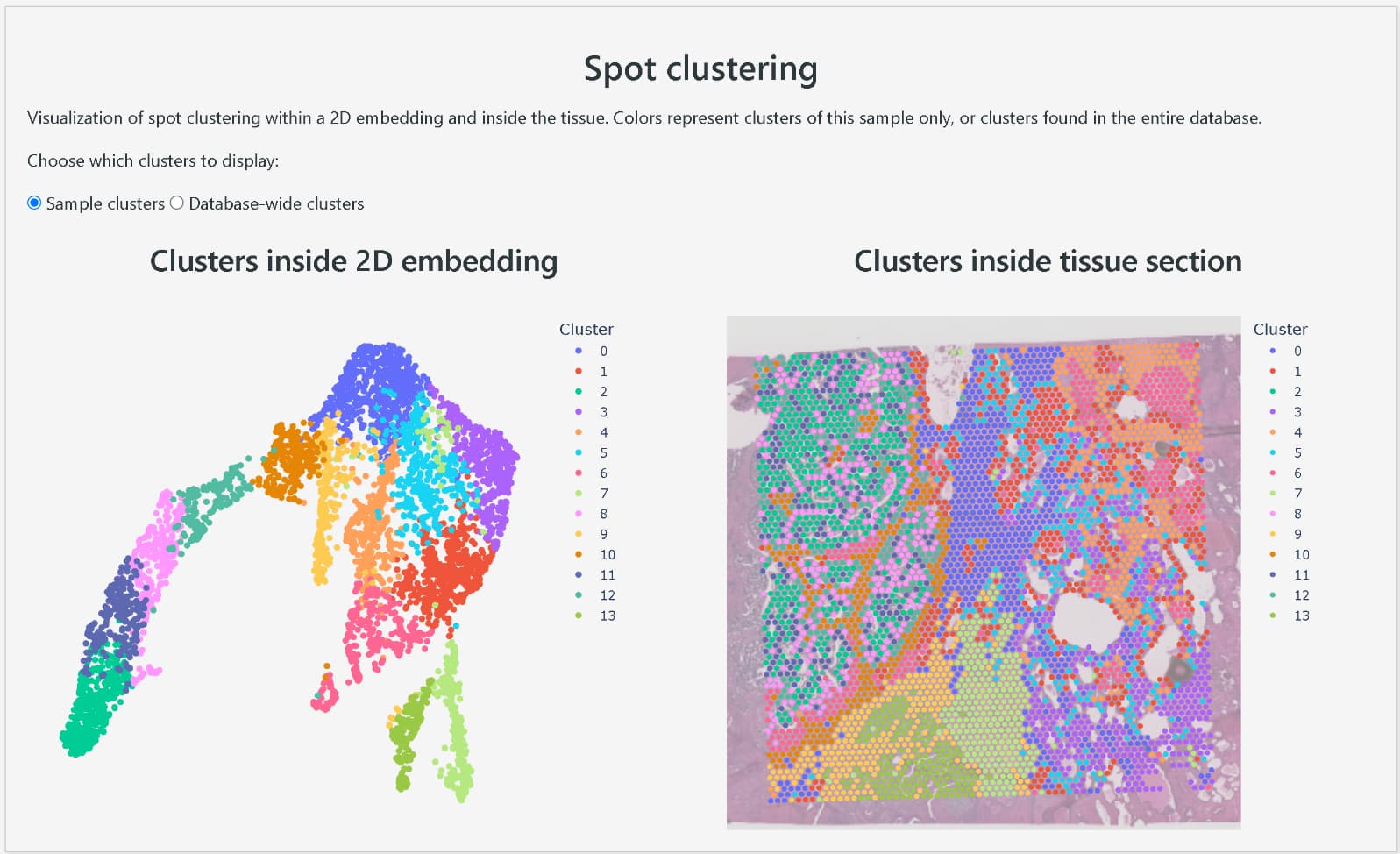
Clustering of each spot based on expression profile
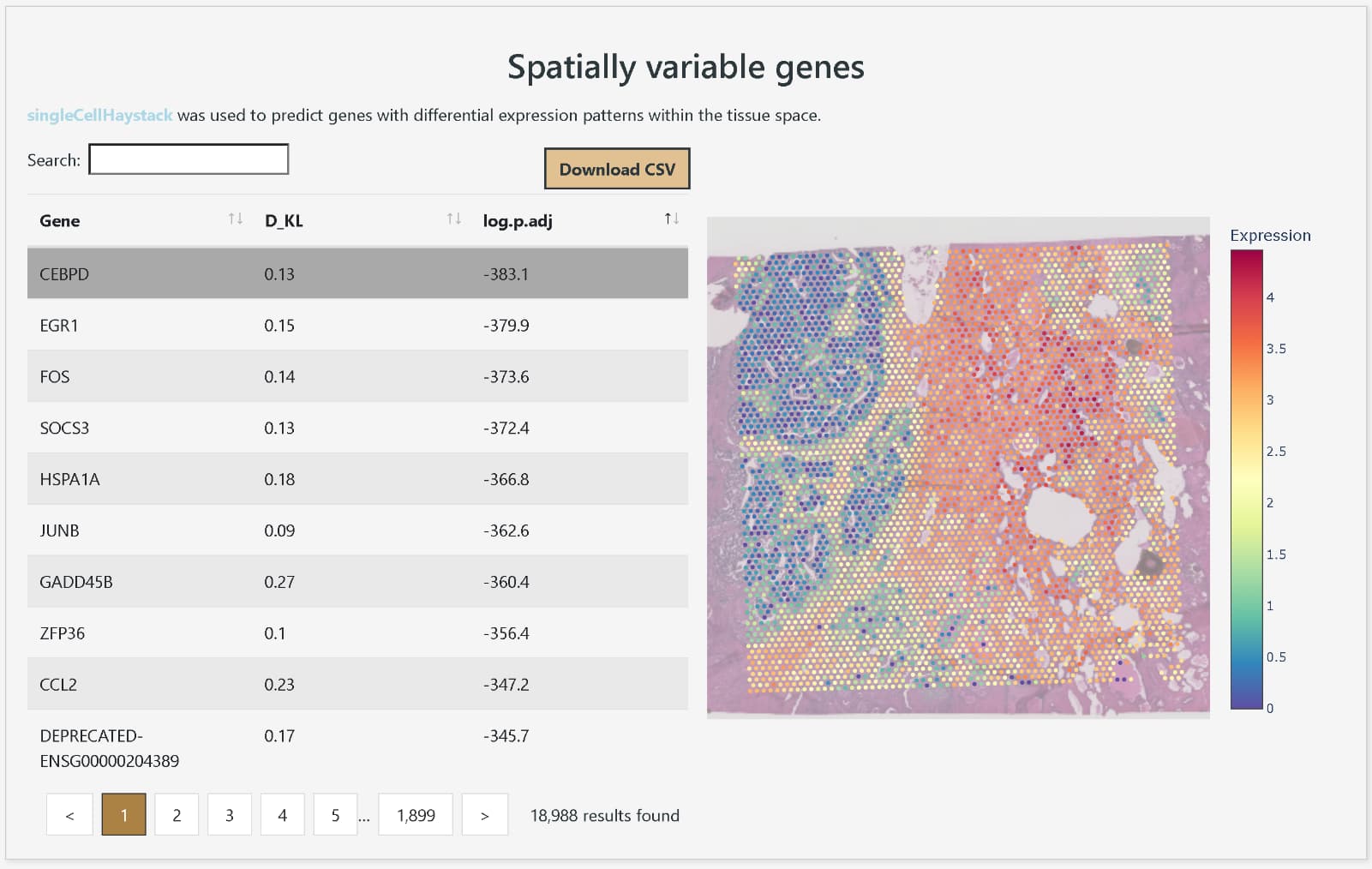
Display of the spatial expression distribution of any gene
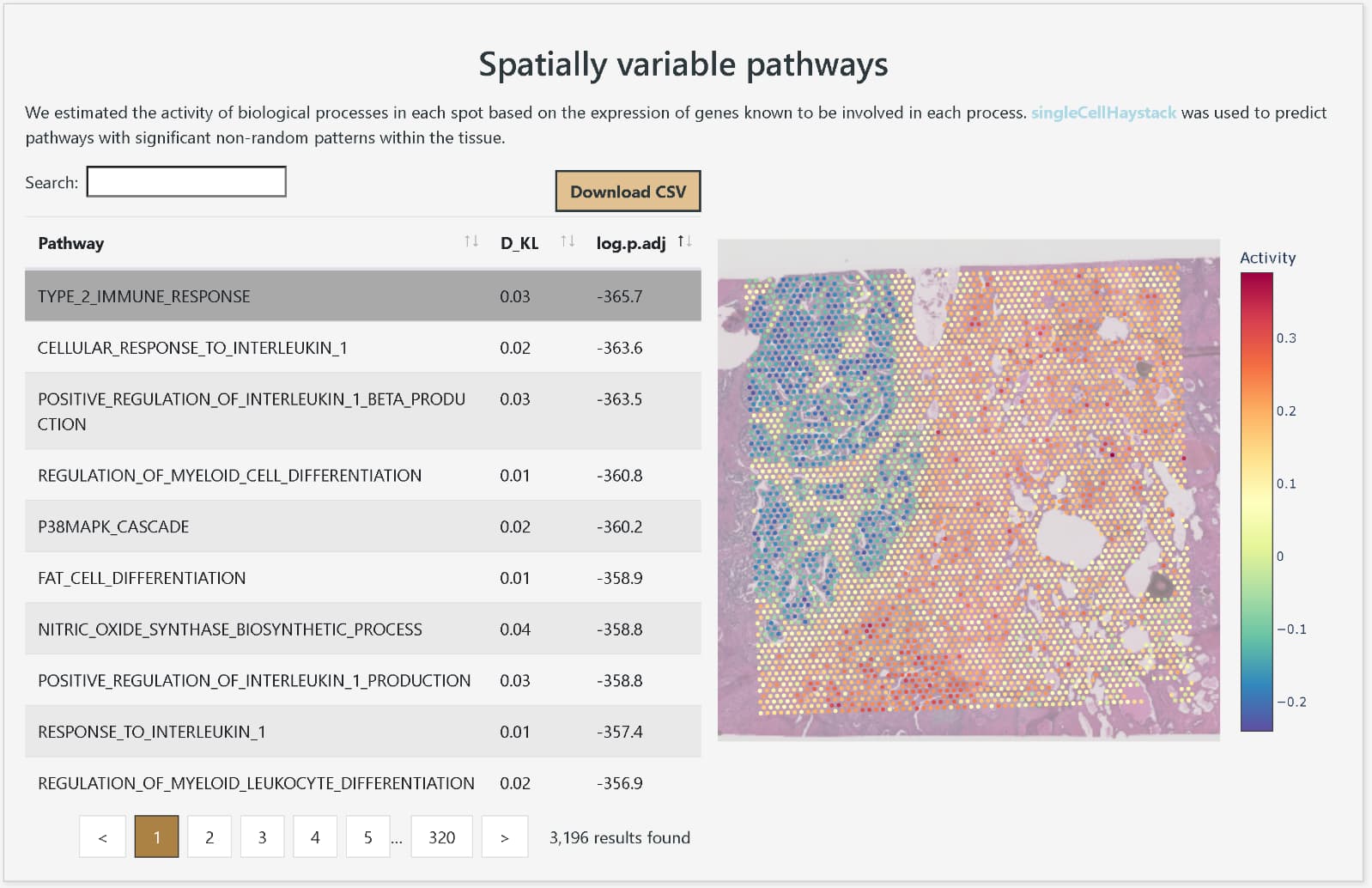
Display of pathways with spatially variable expression
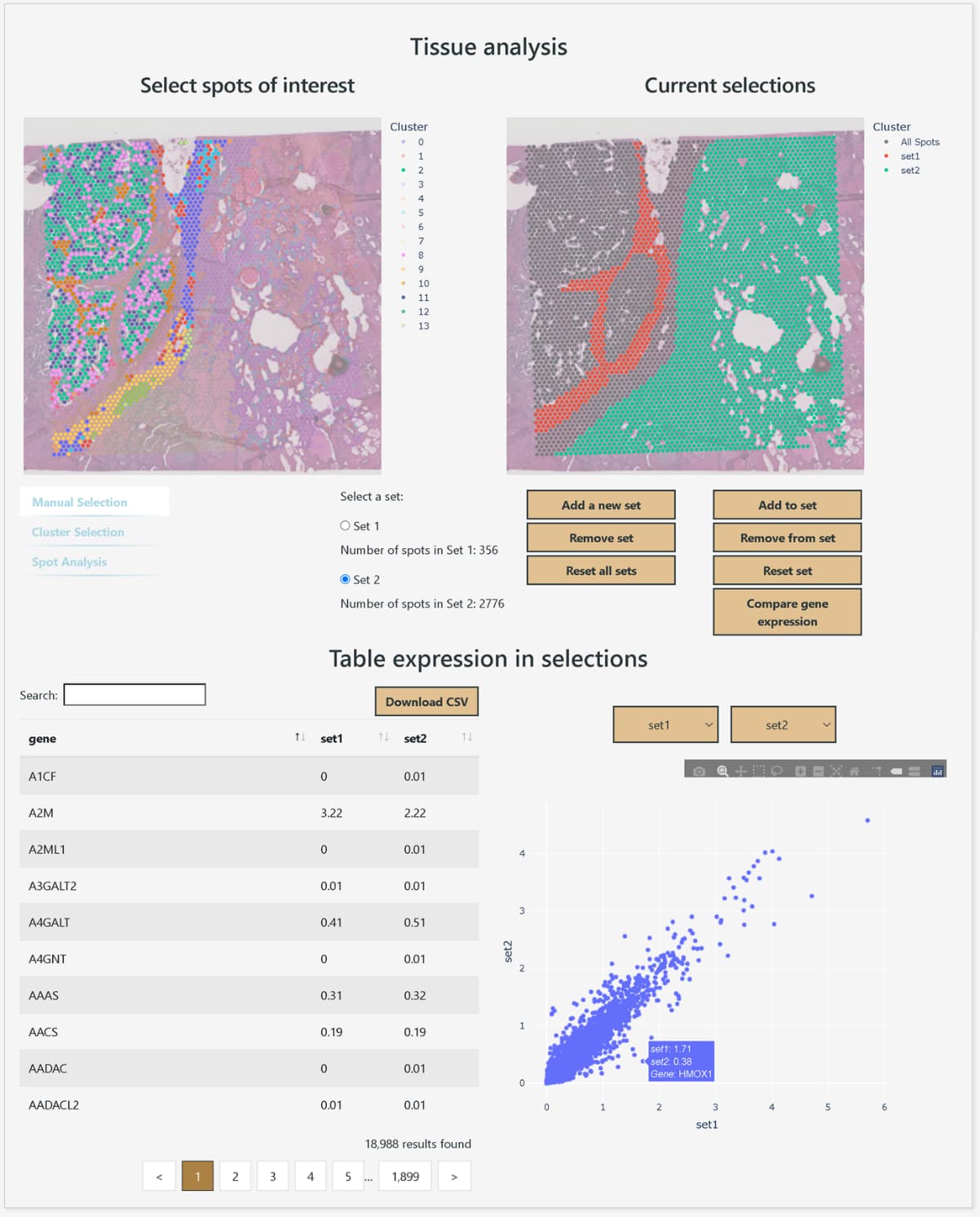
Gene expression comparison between arbitrarily selected areas
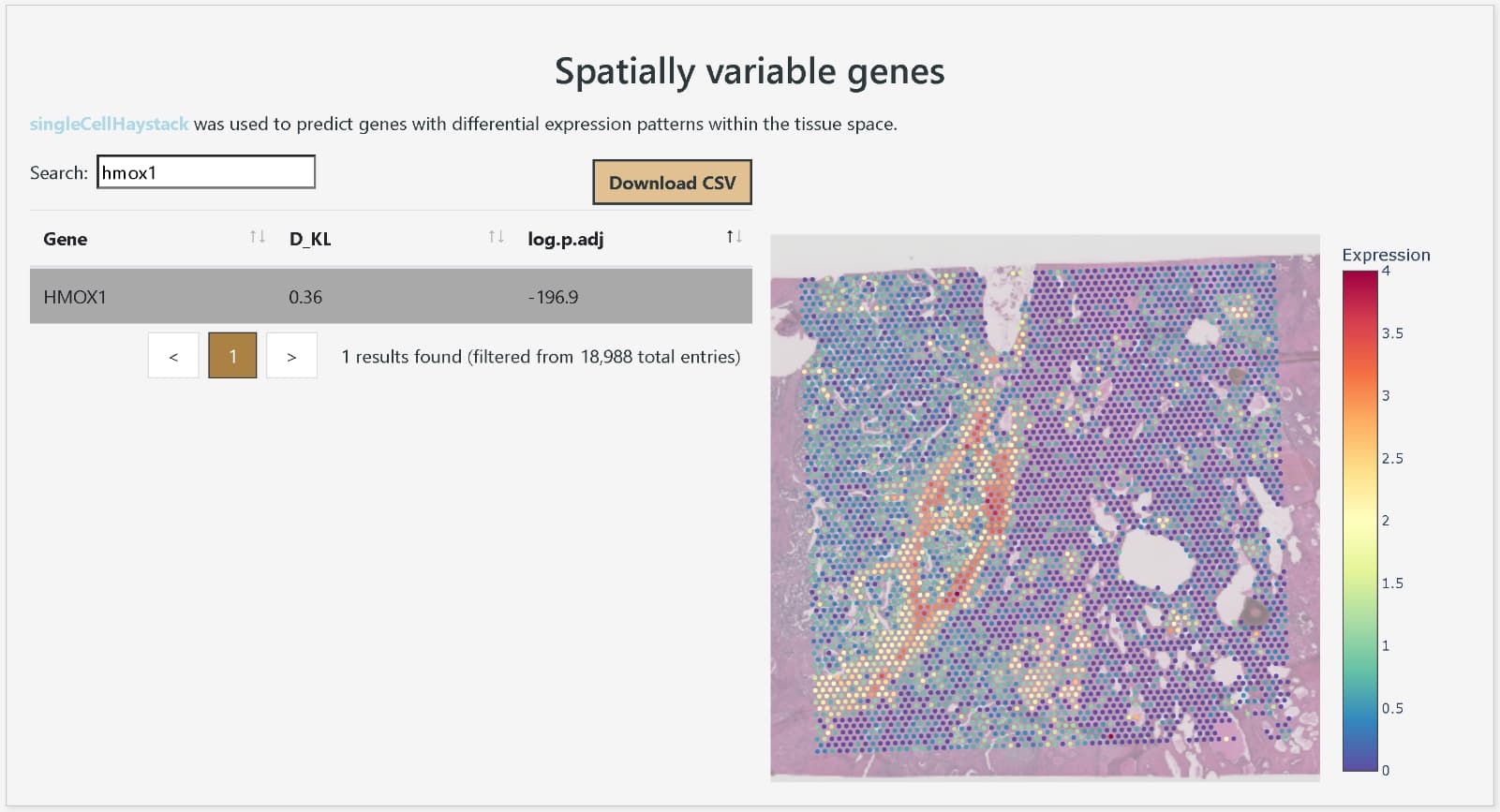
Confirming the high expression of HMOX1 in the tissue surrounding the tumor
Related Links