Traits predicted using Bac2Feature have been added to the Microbiome Datahub
- Others
- Funding
- Database Integration Coordination Program
Dr. MATSUI Motomu at Kyoto University Institute for Chemical Research and his colleagues, published "Bac2Feature", a tool for predicting microbial traits and characteristics from the genome sequences in the scientific journal Bioinformatics Advances on Jun 14, 2025. Along with this, information predicted using Bac2Feature of isolated bacterial genomes and Metagenome-Assembled Genomes (MAGs)【Supplementary Explanation】 has been added to a microbial genome database, Microbiome Datahub.
Dr. MATSUI and his colleagues integrated various prediction methods into a standardized dataset of microbial characteristics and traits, and developed Bac2Feature, which predicts various characteristics and properties of microorganisms from 16S rRNA gene sequences, including cell morphology, Gram staining properties, spore formation ability, motility, genomic information, and environmental characteristics such as growth conditions. According to the paper, Bac2Feature enables research approaches based on microbial characteristics and traits, expanding the potential for applications in medical research and other fields.
On the other hand, Microbiome Datahub developed by Associate Professor MORI Hiroshi at National Institute of Genetics and his colleagues, contains more than 26,000 isolated bacterial genomes and more than 210,000 MAGs identified from metagenomes of various samples. Microbiome Datahub has also released information on the characteristics and traits of microorganisms derived from isolated bacteria and MAGs predicted by Bac2Feature. Since MAGs often do not contain 16S rRNA gene sequences, information on the phylogenetic names of MAGs is used to identify the characteristics and traits of microorganisms. MAGs often includes a large number of genome sequences derived from unknown microorganisms that cannot be cultured in laboratory settings. Dr. MORI states that the information predicted by Bac2Feature available in Microbiome Datahub, enabling researchers to estimate the characteristics and traits of the microorganisms from which a virtual genome MAG was originated, and that is expected to greatly advance MAG research and its practical applications.
For details on Bac2Feature, please refer to the paper "Bac2Feature: an easy-to-use interface to predict prokaryotic traits from 16S rRNA gene sequences."
Bac2Feature was developed by Dr. MATSUI Motomu and his colleagues as part of JST Database Integration Coordination Program (DICP), "Development of an integrated microbiome data hub for microbiome research" (Principal Investigator: Associate Professor MORI Hiroshi, National Institute of Genetics).
Number of data entries in Microbiome Datahub (Jun. 5, 2025)
- Isolated bacterial genomes: 26,076 entries
- Metagenome-assembled genomes (MAGs): 218,653 entries
[Supplementary explanation]
Metagenome-Assembled Genome (MAG): Genome sequences comprehensively analyzed without isolating and culturing microorganisms from soil, environmental water, feces, skin tissue, etc. are called "metagenomes." Metagenomes contain fragmented genome sequences derived from a variety of microbial species that cannot be cultured in a laboratory. By assembling these fragmented sequences on a computer to reconstruct the original microbial genome, the resulting virtual sequence is referred to as a "Metagenome-Assembled Genome (MAG)." MAGs are expected to reveal previously unknown microorganisms, as well as contain useful information for industrial applications, such as novel enzyme genes possessed by these microorganisms.
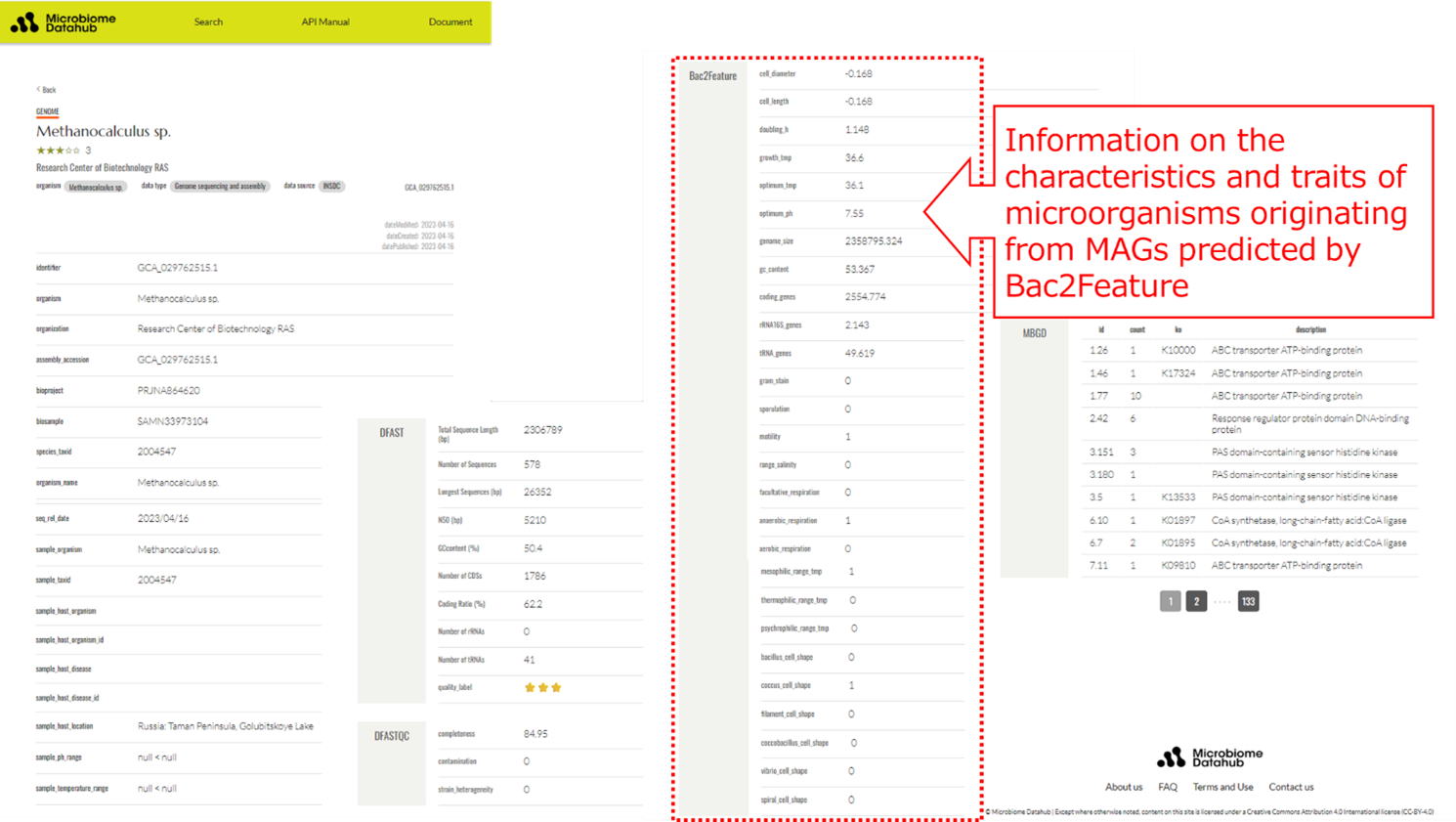
Figure 1: An example of information on the characteristics and traits of Metagenome-Assembled Genomes (MAGs) predicted by Bac2Feature displayed on Microbiome Datahub.
Related Links
- Paper "Bac2Feature: an easy-to-use interface to predict prokaryotic traits from 16S rRNA gene sequences." | Bioinformatics Advances
- Bac2Feature | Command Line Interface / Web Interface
- Microbial genome database, Microbiome Datahub
- Development of an integrated microbiome data hub for microbiome research | NBDC
Outline of the project, etc.