A PDBj paper reporting on the development of an integrated portal for 3D protein structures and genetic mutations, etc. has been published
- Others
- Funding
- Database Integration Coordination Program
A research group of Professor Genji Kurisu and his colleagues at the Institute for Protein Research, Osaka University published a paper on the update of the Protein Data Bank of Japan (PDBj) in the journal "Protein Science" on Feb 20, 2025.
Research groups around the world have determined the protein structures of a large number of proteins from a wide variety of species. The structure data are registered in the Protein Data Bank (PDB) and made publicly available to the entire world. PDBj has registered and processed all protein structure data analyzed in Asia, and has made all PDB data publicly available in cooperation with its bases in the United States and Europe. PDBj also develops original services and tools for searching, visualizing and analyzing 3D protein structure data.
One of the updates reported in this paper is "UniProt Integrated Portal", which was released in Nov 2024. This portal site provides the 3D structure data of PDB for each amino acid sequence (UniProt ID) registered in UniProt, gene mutation information derived from jMorp (Japanese Multi Omics Reference Pannel) and MGeND (Medical Genomics Japan Variant Database) , and interaction information all on one page, allowing comprehensive viewing of information on the 3D structure of any protein. Each 3D structure data displayed on this page is assigned a unique quality evaluation score, which can be used as a reference when selecting which structure data to use. In addition, gene mutation information is displayed in the amino acid sequence viewer, and by selecting any amino acid residue, its position can be confirmed in the 3D structure viewer. This can be useful, for example, in estimating the effect of disease-related gene mutations on protein function.
This paper also reports on "Sequence Navigator Pro ", which allows amino acid sequence similarity searches against UniProtKB/Swiss-Prot and AlphaFold DB data in addition to PDB, "BSM-Arc (Biological Structural Model Archive) ", an archive that accepts computational data based on molecular dynamics, deep learning, etc., and "XRDa (Xtal Raw Data Archive) ", an archive of diffraction images obtained by X-ray diffraction, etc.
PDBj is being developed as a part of the JST Database Integration Coordination Program (DICP), under the project title "Advanced development of Protein Data Bank extended as the Data-driven research platform ".
For more detail, please see a paper "Protein Data Bank Japan: Improved tools for sequence-oriented analysis of protein structures"
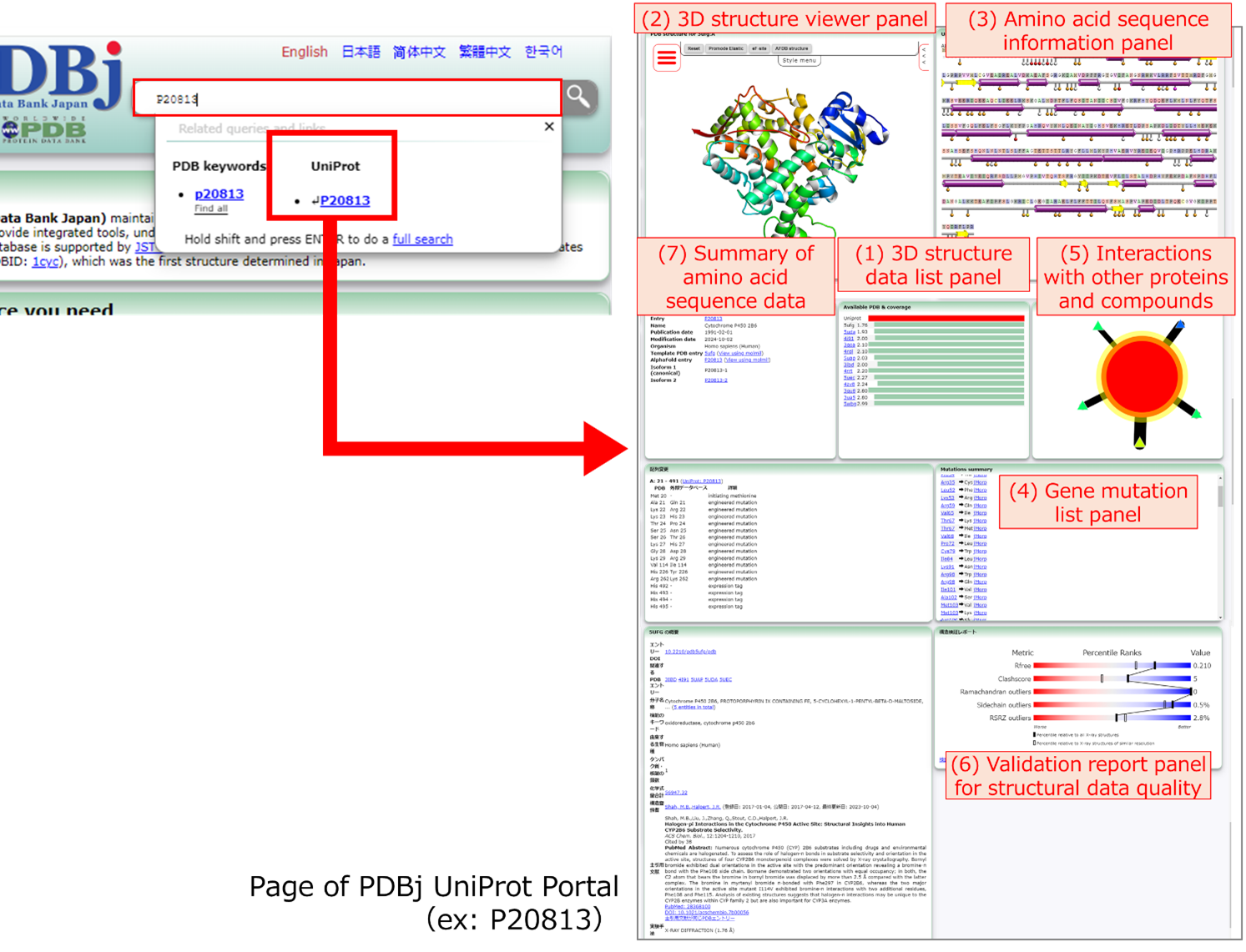
Figure 1: The new portal page can be accessed by entering a UniProt ID in the search window of the PDBj top page . In addition, links to the UniProt entry pages are provided from the corresponding PDB entries. For each UniProt ID, a page is available that contains various information related to that UniProt ID and integrates data from various secondary databases developed by PDBj.
(1) The 3D structure data list panel displays a list of PDB IDs associated with the UniProt ID and the quality evaluation score of each entry. As shown in Figure 2, by clicking on any PDB ID, the data associated with that 3D structure entry can be viewed. (2) The 3D structure viewer panel allows viewing of the 3D structure from the PDB (see also Figure 4) using Molmil, the original molecular viewer developed by PDBj. (3) The amino acid sequence information panel displays data derived from UniProt and the genetic mutation information obtained from jMorp. (4) The gene mutation list panel lists the gene mutations of the relevant protein, with links to the respective detailed pages of jMorp. In addition, (5) information on interactions with other proteins and compounds in the selected protein structure is shown, and (6) the validation report to evaluate the quality of the structure data can also be viewed in one place.
This figure shows the page for UniProtID:P20813 (https://pdbj.org/uniprot/P20813/) as an example. When this page is accessed, the data for PDB ID:5ufg is displayed by default. Therefore, the same information is displayed for both "https://pdbj.org/uniprot/P20813/" and "https://pdbj.org/uniprot/P20813/5ufg/".
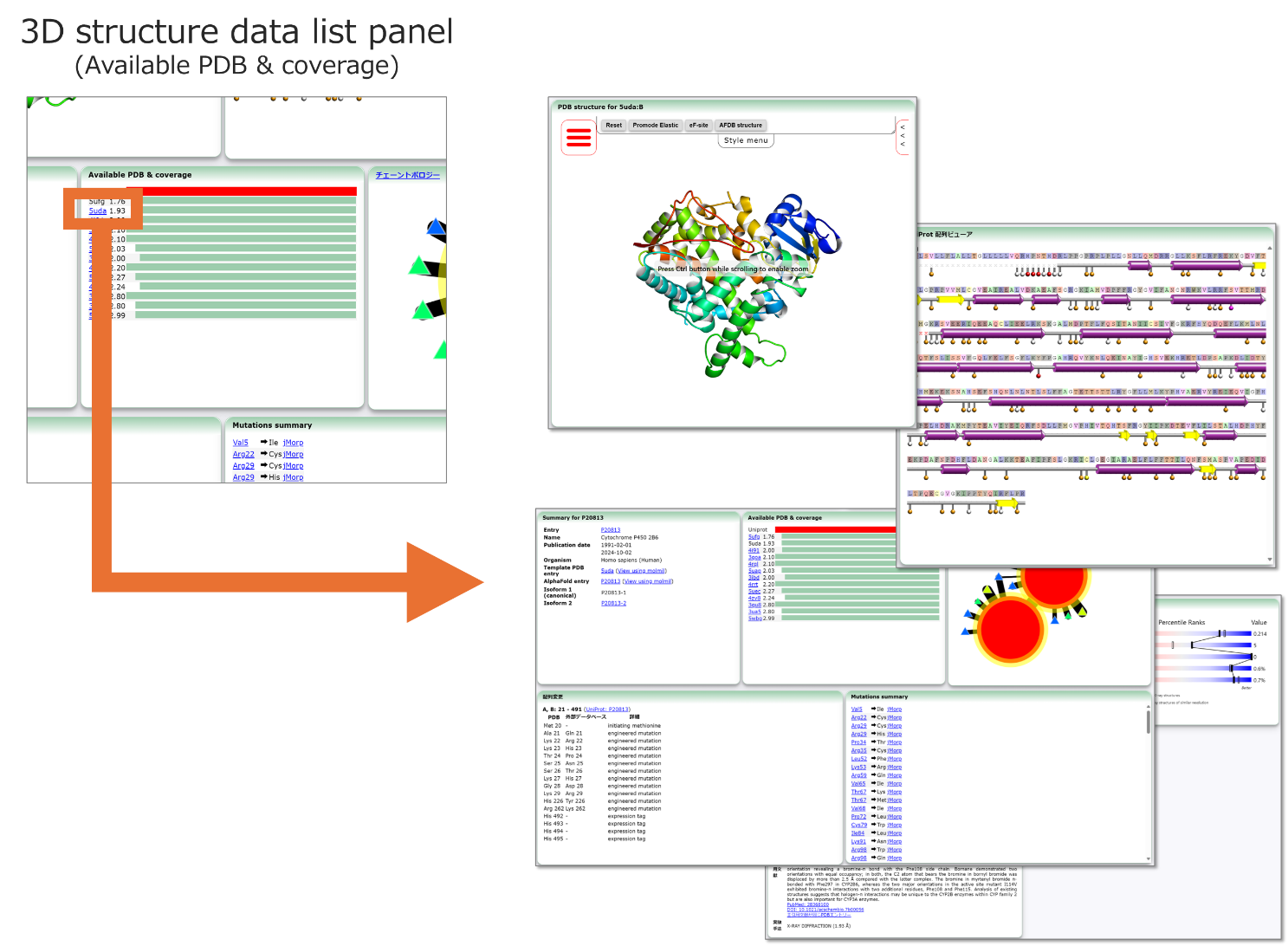
Figure 2: Clicking on any 3D structure data in the 3D Structure Data list panel displays the data associated with that 3D structure entry. A quality evaluation score is given for each 3D structure. This score is calculated based on the resolution as well as the percentage of the full-length sequence for which 3D structure information is available for each given entry.
This figure shows the case of selecting PDB ID:5uda. Starting from "https://pdbj.org/uniprot/P20813/" and selecting 5uda from the list, the page "https://pdbj.org/uniprot/P20813/5uda" is loaded.
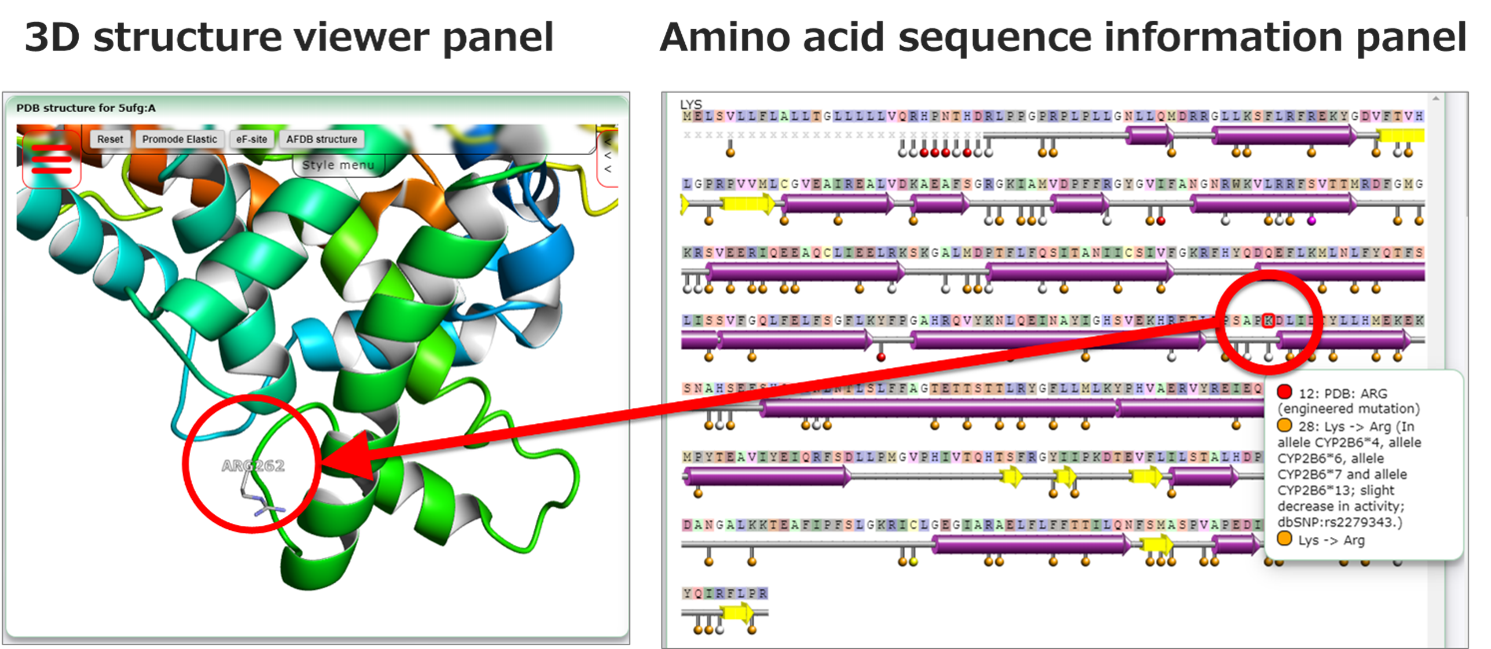
Figure 3: Clicking on any amino acid residue in the amino acid sequence data panel will display the position of the amino acid residue in the structure in the 3D structure viewer panel. As an example, this figure shows the effect of clicking on ARG262 with PDB ID:5ufg is shown (https://pdbj.org/uniprot/P20813/5ufg?select=262).
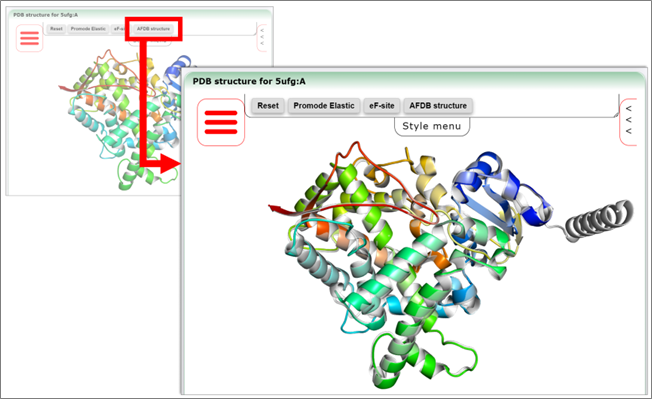
Figure 4: The 3D structure viewer panel can display 3D structures in various formats, including ribbon models, and can overlay Promode Elastic, eF-site and Dynamics DB secondary database data developed and provided by PDBj. AlphaFold predictive structure data can also be displayed when available ("AlphaFold DB " is a web service developed and provided by EBI Europe in collaboration with Google DeepMind).
Related links